|
I recently earned my Ph.D. degree in Computer Science from Zhejiang University in June 2024, under the supervision of Professors Huajun Chen, Ningyu Zhang and Xiaohui Fan. I have joined the National Institutes of Health (NIH) as a postdoctoral fellow, with Dr. Zhiyong Lu as my mentor. My research primarily focuses on AI4Bioinformatics. I am interested in using large language models and graph neural networks to analyze biomedical data, e.g. molecules, proteins, and single-cell data. Feel free to reach me if you are interested in my research! Email / Google Scholar / DBLP / Github / Twitter |
|
|
|
|
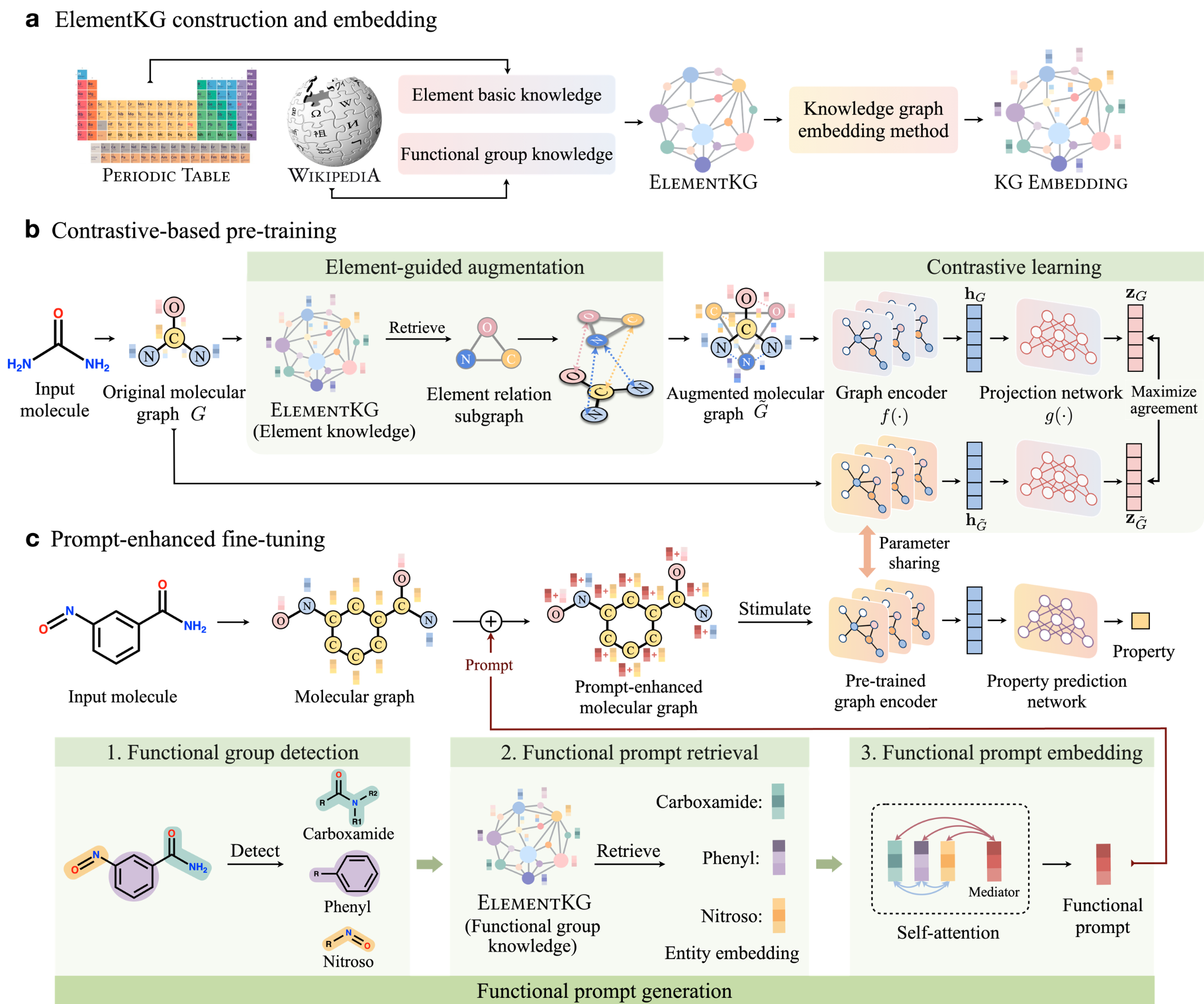
Yin Fang, Qiang Zhang, Ningyu Zhang, Zhuo Chen, Xiang Zhuang, Xin Shao, Xiaohui Fan, Huajun Chen Nature Machine Intelligence (IF=25.90), 2023 Project page / Paper KANO is a Knowledge graph-enhanced molecular contrAstive learning framework with fuNctional prOmpt, designed to exploit fundamental domain knowledge in both pre-training and fine-tuning phases. |
|
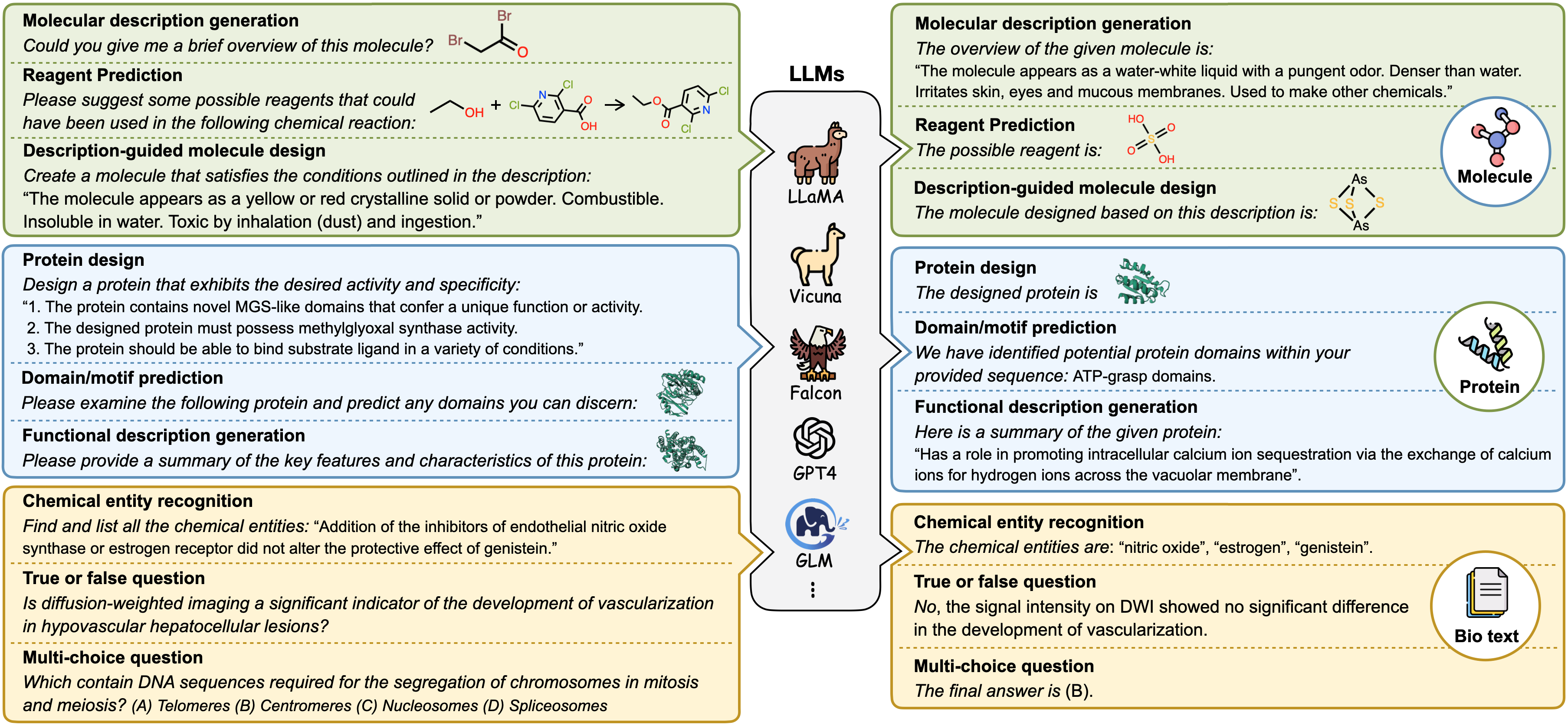
Yin Fang, Xiaozhuan Liang, Ningyu Zhang, Kangwei Liu, Rui Huang, Zhuo Chen, Xiaohui Fan, Huajun Chen ICLR, 2024 Project page / Paper Mol-Instructions is a Large-Scale Biomolecules Instruction Dataset for Large Language Models. |
|
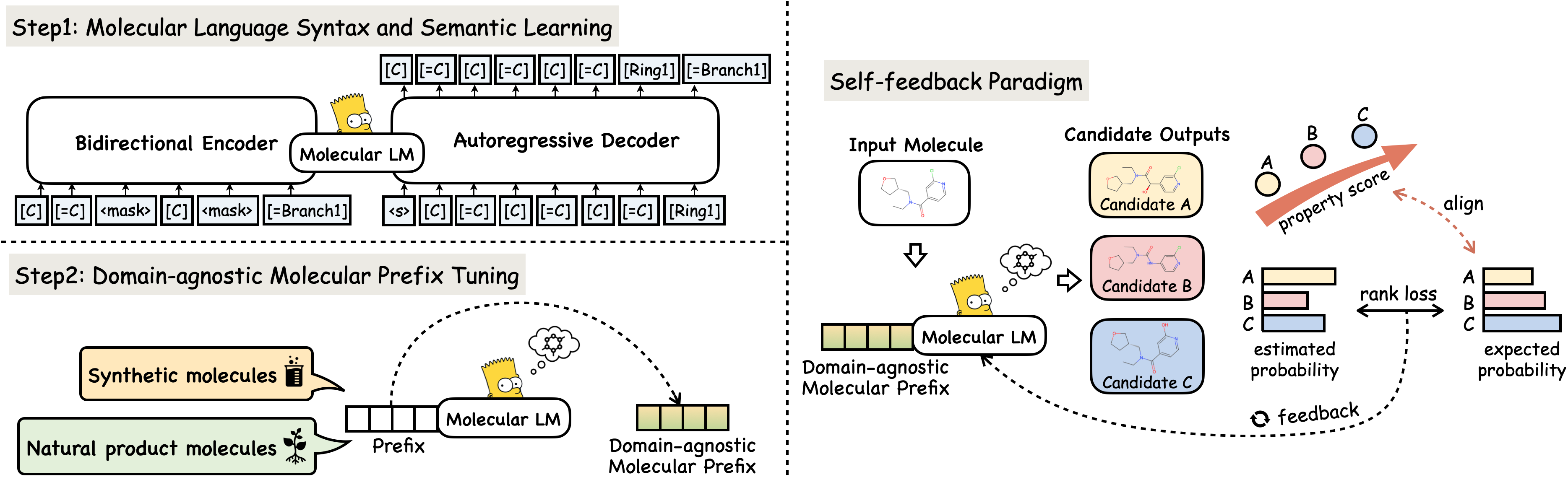
Yin Fang, Ningyu Zhang, Zhuo Chen, Lingbing Guo, Xiaohui Fan, Huajun Chen ICLR, 2024 Project page / Paper MolGen is a pre-trained molecular language model tailored specifically for molecule generation. |
|
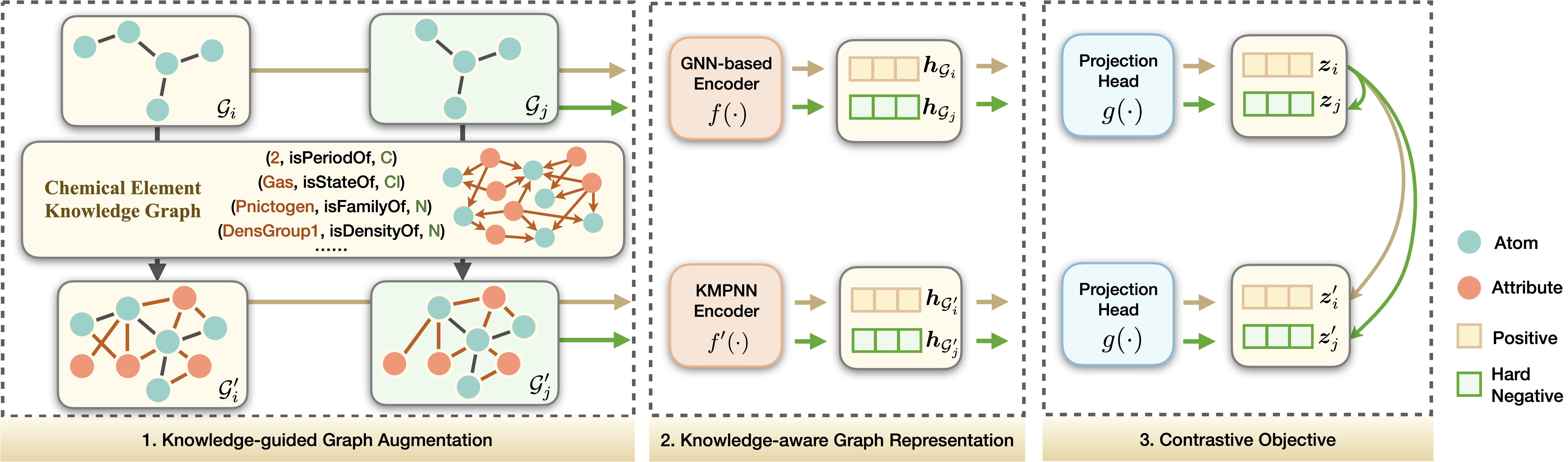
Yin Fang, Qiang Zhang, Haihong Yang, Xiang Zhuang, Shumin Deng, Wen Zhang, Ming Qin, Zhuo Chen, Xiaohui Fan, Huajun Chen AAAI, 2022 Project page / Paper KCL is a novel Knowledge-enhanced Contrastive Learning framework for molecular representation learning, built upon a Chemical Element Knowledge Graph (KG) that summarizes microscopic associations between elements. |
|
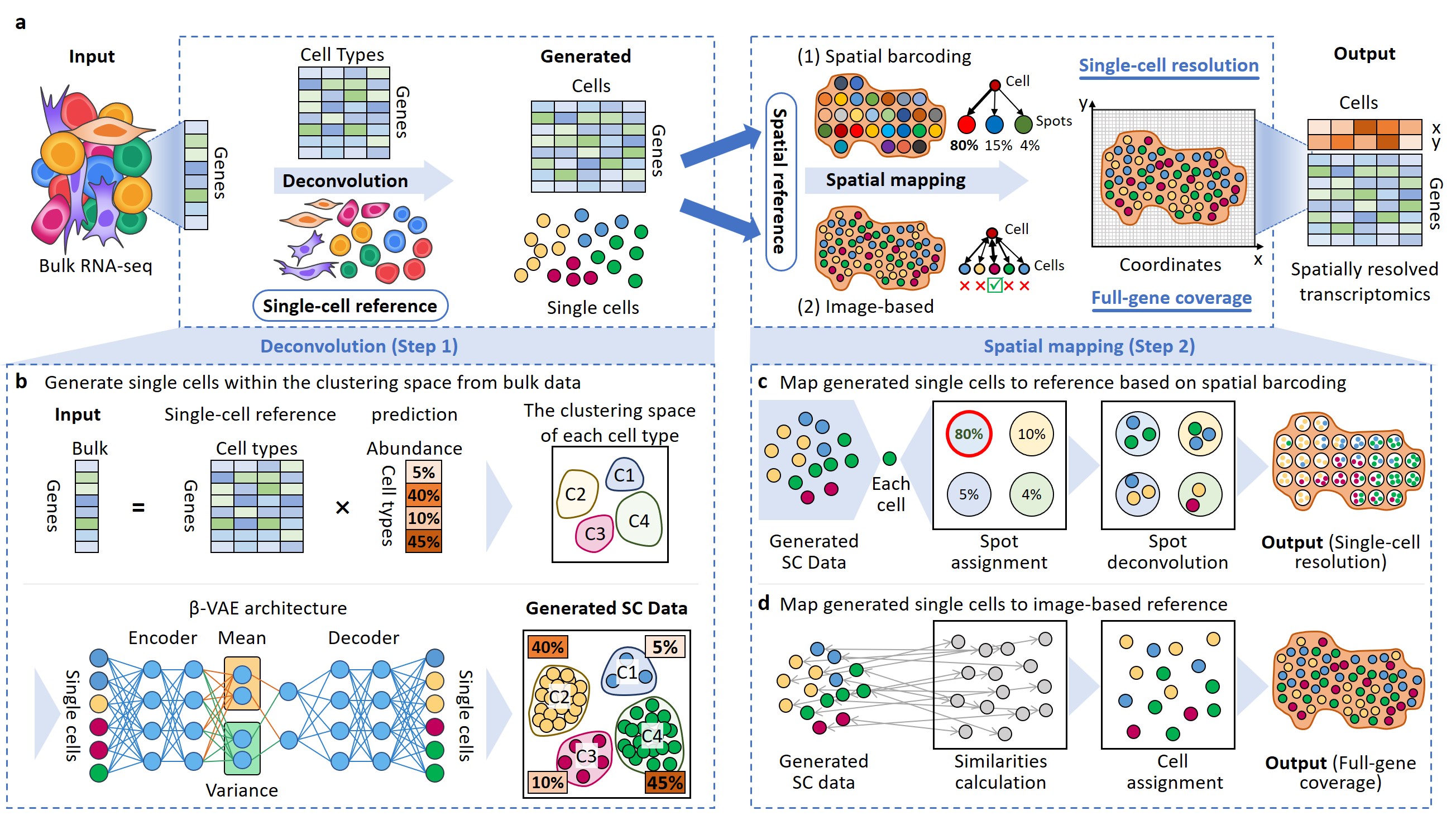
Jie Liao*, Jingyang Qian*, Yin Fang*, Zhuo Chen*, Xiang Zhuang*, Ningyu Zhang, Xin Shao, Yining Hu, Penghui Yang, Junyun Cheng, Yang Hu, Lingqi Yu, Haihong Yang, Jinlu Zhang, Xiaoyan Lu, Li Shao, Dan Wu, Yue Gao, Huajun Chen, Xiaohui Fan Nature Communications (Editors' Highlights, Top 25 in 2022) (IF=17.69), 2022 Project page / Paper Bulk2Space is a two-step spatial deconvolution method based on deep learning frameworks, which converts bulk transcriptomes into spatially resolved single-cell expression profiles. |
|
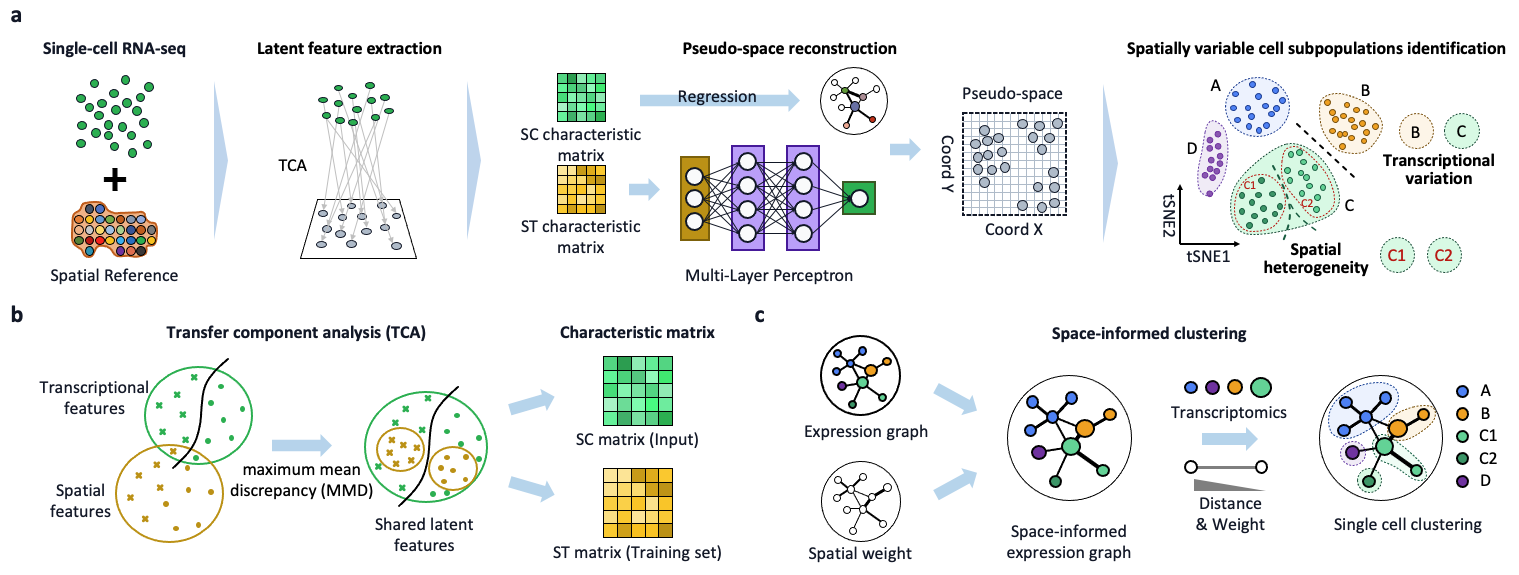
Jingyang Qian*, Jie Liao*, Ziqi Liu*, Ying Chi*, Yin Fang, Yanrong Zheng, Xin Shao, Bingqi Liu, Yongjin Cui, Wenbo Guo, Yining Hu, Hudong Bao, Penghui Yang, Qian Chen, Mingxiao Li, Bing Zhang, Xiaohui Fan Nature Communications (Editors' Highlights) (IF=17.69), 2023 Project page / Paper scSpace (single-cell and spatial position associated co-embeddings) is an integrative algorithm that integrates spatial transcriptome data to reconstruct spatial associations of single cells within scRNA-seq data. |
|
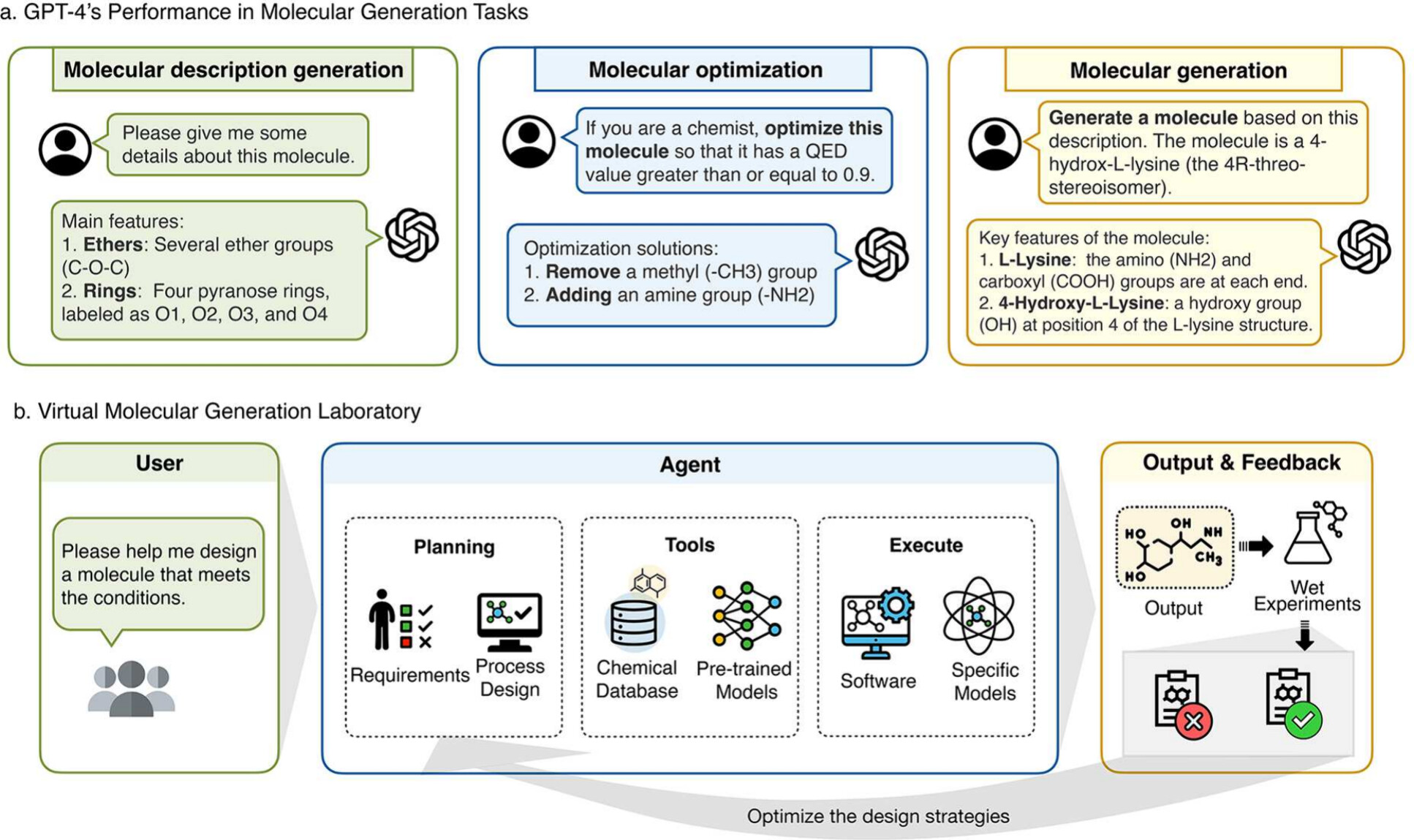
Jinlu Zhang, Yin Fang, Xin Shao, Huajun Chen, Ningyu Zhang, Xiaohui Fan Journal of Chemical Information and Modeling (IF=6.16), 2023 Paper This paper proposes possible directions for future molecular science research. These suggestions aim to forge new paths for exploring the intricacies of molecular structures, potentially bringing new efficiencies and innovations in the field. |
|
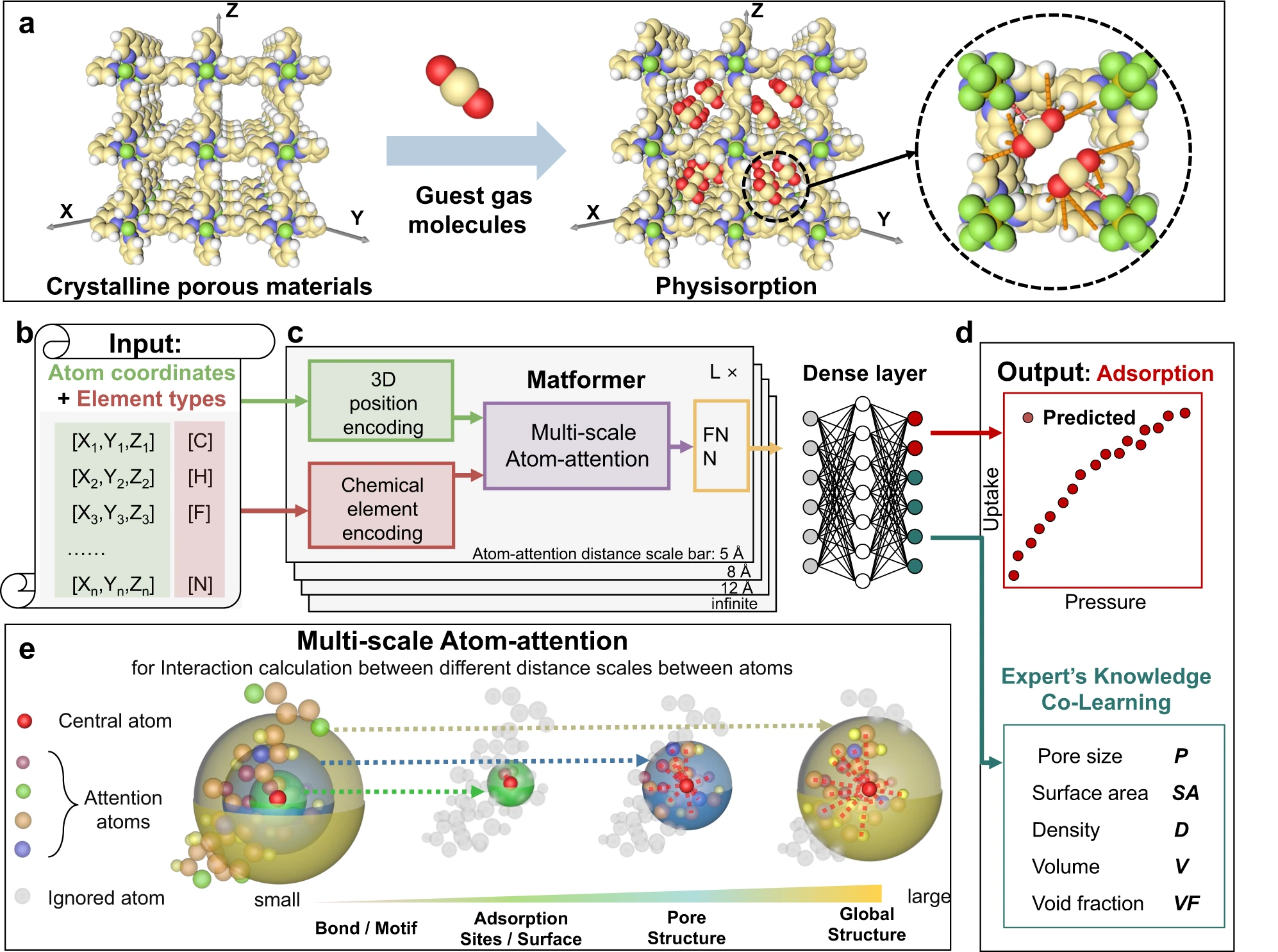
Jiyu Cui, Fang Wu, Wen Zhang, Lifeng Yang, Jianbo Hu, Yin Fang, Peng Ye, Qiang Zhang, Xian Suo, Yiming Mo, Xili Cui, Huajun Chen, Huabin Xing Nature Communications (IF=17.69), 2023 Project page / Paper DeepSorption is a spatial atom interaction learning network that realizes accurate, fast, and direct structure-adsorption prediction with only information of atomic coordinate and chemical element types. |
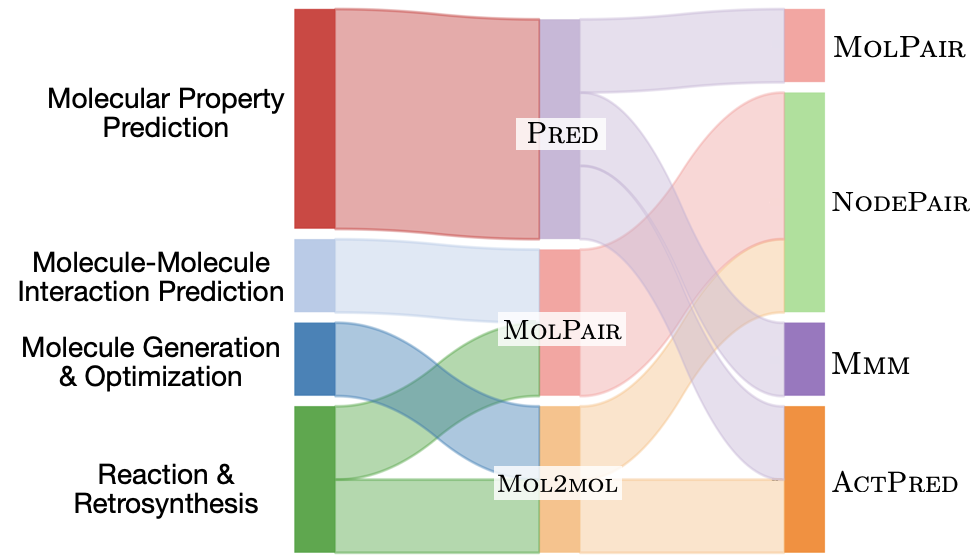
|
Yin Fang, Qiang Zhang, Zhuo Chen, Xiaohui Fan, Huajun Chen KSEM, 2024 arXiv This paper offers a comprehensive review of molecular learning, with a focus on the knowledge-informed paradigm transfer approach. It provides an overview of the various paradigms and their technical solutions, as well as a summary of the external domain knowledge used to guide the transfer process for each molecular learning task. |